Computational evolution of development in plants
Keywords
evolution, development, computational models, plants, gene regulatory networks
Research interests
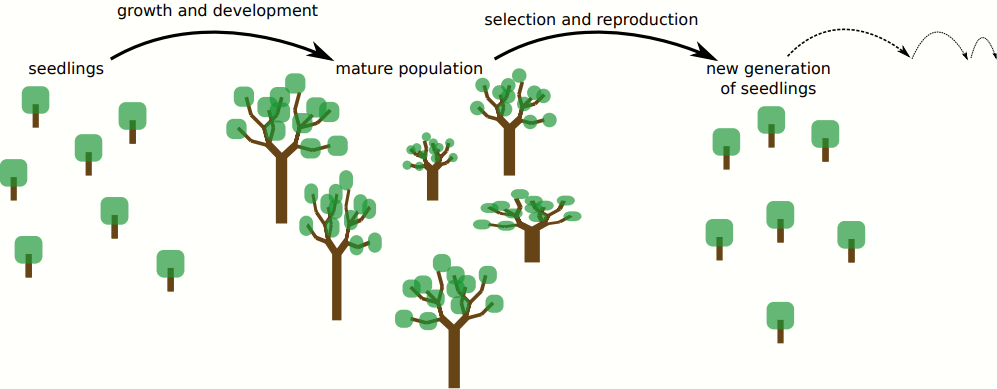
At the SLCU I work on these research interests by studying the evolution of plant organs, in particular the apical stem cell niche. I hope to find general principles of evolution that are shared between plants and animals, as well as interesting and informative differences. In both cases, embryonic development generates a complete multicellular organism from a single, fertilized egg cell; it's a program which is (mostly) encoded in the genome, but then requires coordinated interactions between several different levels of organisation. Because of this complex mapping from the genotype to the phenotype, mutations can have wildly different effects on the developmental process, ranging from no effect to lethality. We therefore need to understand how development influences the effects of mutations and how mutations in turn alter the developmental program, to understand the evolution of multicellular organisms as a whole. The time scales of these evolutionary processes are vast, and therefore difficult to study in the lab alone. I therefore construct computer models in which I simulate an evolving population of plants which have to grow and develop. When a successful plant reproduces, its offspring will inherit their genome, with mutations. In this way, I can simulate thousands of years of evolution in days or weeks, and store a perfect fossil record of all the changes in the developmental program. This allows me to study in detail how, over evolutionary time, the accumulation of mutations leads to new developmental programs that make new organs. By switching up the starting conditions
