Submitted by Administrator on Tue, 08/12/2020 - 16:05
The Prabakaran group has identified, cataloged, and classified protein translations from noncoding genomic regions of multiple organisms. Their work, which is to appear in three separate publications (see below), shows that these ‘novel’ genomic regions cannot be defined by our current understanding or definition of a gene. Hence, they call these novel regions as novel Open Reading Frames or as nORFs. The three papers discuss in detail the role of these nORFs, their disruption in diseases, their evolution, and how they may re-define our fundamental understanding of genes and evolution.
In Puntambekar, S. et al., (Evolutionary divergence of novel open reading frames in cichlids speciation. To appear in Scientific Reports on Dec 9th) they show evidence for a strong correlation between the time-scales of the adaptive radiation of two cichlid fishes and the emergence of species-specific nORFs. Although these species-specific nORFs make both transcripts and proteins, it is not clear what the role of these novel proteins might be. They hypothesize that the novel genes and proteins are essentially minted from noncoding regions, and they are kept (fixed in a population) if they serve a purpose.
In Neville, M et al., (A platform for curated products from novel open reading frames (nORFs) prompts reinterpretation of disease variants.) they have cataloged, curated, annotated, and released ~194,000 nORFs in the human genome (nORFs.org). They have performed population genetic - based global analysis on the heritability and negative selection of mutations on these nORFs. They show that the mutations in nORFs do have physiological consequences, are deleterious, and are selected out. They also show that majority of mutations that are often annotated as benign or variant of unknown significance have to be re-interpreted based on their observation of the presence nORFs in the vicinity and their assessment that these mutations cause either premature truncation (stop-gain) or elongation (stop-loss) of nORF proteins.
In Erady, C. et al., (Pan-cancer analysis of transcripts encoding novel open reading frames (nORFs) and their potential biological functions.) they show that nORFs are dysregulated (both up- and down-regulation and mutation) in all 33 cancers, some nORF disruptions strongly correlate with the survival of patients. More importantly, they show that nORF proteins can form protein structures, can undergo biochemical regulation like known proteins, and be targeted by drugs in case they are disrupted.
All these observations essentially mean that under stress, or physiological changes, or adaptive radiation nature tinkers with the ‘evolvable’ noncoding genome to make new parts as needed. If this is true, they speculate that all the known protein-coding genes in a population or species are essentially playing the role of house-keeping genes, and the nORFs are the ones that give rise to tissue- or species-specificity. Prabakaran group is excited about these results and is embarking on a journey to identify more nORFs in the human genome with the hope to find whether they are disrupted in diseases and whether they can be used for treating diseases.
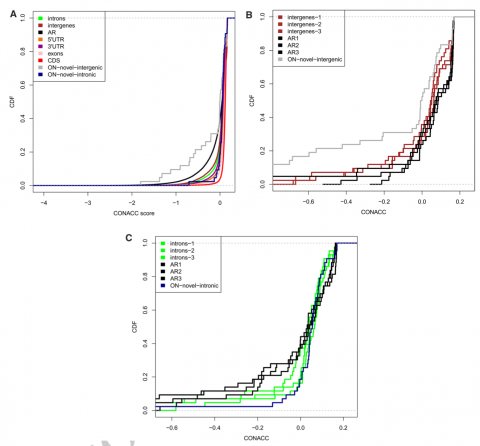
Figure legend: Distribution of conservation-acceleration (CONACC) scores calculated using phyloP for nORFs observed in cichlids fish Oreochromis niloticus (ON)
CONACC scores were computed over all branches of the cichlid’s phylogeny, and used to detect the departure from neutrality in nORF regions and also in the other known annotated features of the genome like CDS, 5′UTR, 3′UTR, introns, intergenes and ancient repeats (AR). The analysis of the cumulative distributions (Fig. A) of the phylop scores of ON’s known annotated features showed that the CDS regions (red line) were most conserved while the AR’s were least conserved. This is intuitive as the functional coding regions are expected to have more evolutionary constraints than the non-functional repeat regions. The distribution of CONACC scores of all the annotated features were significantly different than that of AR (Welch t-test, p-value < 0.05) (Fig. A). Conservation scores were also mapped to the 9 ON novel intergenic and 27 ON’s novel intronic regions. As these novel regions are very few compared to the AR, we sampled 10,000 times, from all the AR regions, to randomly pick one length-matched AR per nORF transcript. The distribution of CONACC scores for these length-matched, equal sample-sized AR regions were significantly different (Welch t-test, p-value < 0.05) than the novel intergenic regions (Fig. B) for 7519/10,000 times; and only 2338/10,000 times for the novel intronic regions (Fig. C). Compared to AR, the 9 novel-intergenic regions in ON showed a shift towards more accelerated CONACC scores (gray line in the graph), whereas the 27 novel-intronic regions showed a non-neutral substitution rate with shift towards more conserved CONACC scores (blue line in the graph). This indicates that these regions which are varied in all the cichlids, might contribute to the phenotypic variation in ON.
