MIMIC Lab
Molecular Innovations in Microbial Interactions and Chromatin
Keywords:
phylogenetics; genomics; synthetic biology; genetic conflicts
Research Interests:
DNA genomes have been adopted by almost all forms of life. Consequently, the protein machinery that cells need to use their genetic information is both essential and largely well conserved. Beyond its sequence, DNA doesn’t differ much between altogether unrelated species, and for this reason DNA binding proteins often share structural similarities. The universality of DNA and DNA binding proteins is what makes horizontal gene transfer and synthetic biology possible. However, this comes at price, as DNA binding proteins inevitably become a weak node of cellular systems, often prone to interference during genetic conflicts.
The focus of the Hocher group is twofold:
-Understanding and engineering chromatin:
The tools of synthetic biology are now sufficiently mature to be applied beyond the generation of individual genetic circuits. In fact, they allow us to rethink and redesign entire systems of gene expression. Our group seeks to harness and engineer the diverse properties of chromatin. Part of this endeavour is dedicated to discovering chromatin proteins with new properties across the tree of life. We focus on the large-scale epigenetic regulation allowed by chromatin. Combining synthetic biology and chromatin evolution, our goal is to recreate minimal chromatin based epigenetic systems from the ground up.
-Discovering and engineering proteins that mimic DNA:
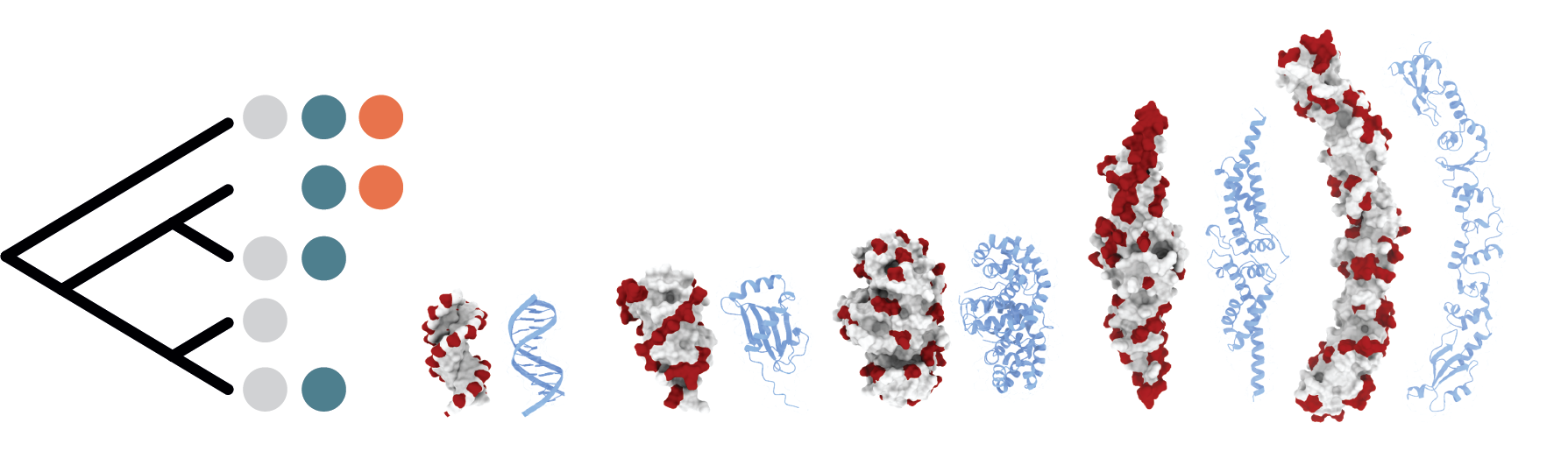
Successful mimicry is a key survival strategy for many plants, insects, and even vertebrates. Mesmerizing camouflage or decoy tactics have evolved multiple times and are among the most intuitive examples of phenotypic convergence. The lab focuses on mimicry at the molecular level. We study proteins that have evolved to mimic DNA. While we know in detail a handful of case of proteins that mimic DNA, fewer than 30 DNA mimicking proteins (DMPs) have been described. Importantly, those that we know of interact with and, in the case of viral-encoded DMPs, disrupt key pathways for bacterial survival, including defence systems against exogenous elements (restriction modification systems, BREX, CRISPR), chromatin organization, and transcription. Studying and engineering this peculiar family of protein has the potential to reveal new way of inhibiting DNA binding proteins, and to advance our understanding of molecular mimicry.
Our lab combines computational structural biology, large-scale phylogenomics, genetics and experimental evolution to study and engineer DNA mimics. The lab uses computational and molecular biology to enable systematic discovery of new DMPs. We couple phylogenomics and directed evolution to design new DNA mimicking proteins and use phylogenomic analysis and genetics to dissect the impact of pathogenic DMPs on bacterial genome function and evolution.
Join Us
We welcome applications from enthusiastic students and talented postdocs. The main requirement is to be creative and rigorous. If you are interested in joining the lab, send an email to Antoine Hocher outlining your research interests and a C.V.
Acknowledgements:
Work in the Hocher lab is made possible thanks to funding of the Wellcome Trust.
