Transposable elements and the evolution of gene regulatory networks
Keywords
Epigenetics, Transposable elements, KRAB-ZFPs, Evolution, Gene expression, Bioinformatics
Research interests
Over evolutionary time, successive waves of transposable elements (TEs) have colonized mammalian genomes, including ours; nowadays, over half of human DNA can be traced back to retroviruses and other mobile elements (LINE, SINE, SVA, DNA transposons). This process is dynamic: as more of these genetic invaders come in, old ones are gradually erased by the accumulation of mutations, or deleted by recombination events. However, some TEs are retained through evolution as they are in some way beneficial; for example, if they integrated near a gene and can affect its expression by providing a new enhancer or an alternative promoter. This process of domestication is a major force shaping genomes and driving evolution of gene regulatory networks.
Our lab is focused on the participation of KRAB zinc finger proteins (KRAB-ZFPs) in this process. KRAB-ZFPs emerged at the dawn of tetrapods, yet constitute the largest subfamily of DNA binding factors in mammals. They are expanding at a dramatic pace, with new members found at every phylogenetic branch; illustrative of this fact, only 80 genes are shared between mouse and human out of their respective 350 KRAB-ZFPs. They primarily target endogenous retrotransposons and act as silencers through the recruitment of TRIM28 and the subsequent deposition of repressive epigenetic marks (H3K9me3 and DNA methylation). KRAB-ZFPs and TEs are thought to be locked in a relentless evolutionary arms race, with new KRAB-ZFPs continuously emerging to tackle the genomic threat of novel invading transposable elements.
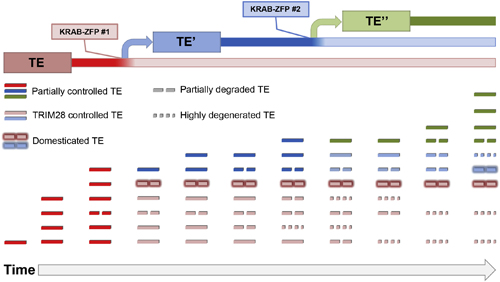
When we revealed the evolutionary interplay between human KRAB-ZFPs and their genomic targets, we found a majority to be old, targeting equally ancient remains of TEs, millions of years after these had lost all replication potential. This is at odds with the main established role of KRAB-ZFPs: the transcriptional control of young and active TEs in embryonic stem cells. Moreover, we found that a large proportion of these largely degraded TEs still contain docking sites for transcription factors and are epigenetically active in certain cellular contexts; therefore, they are potentially domesticated and could participate in the rewiring of transcriptional networks. Furthermore, expression patterns of KRAB-ZFPs are quite diverse and not restricted to embryonic stem cells, hinting at roles outside the repression of TEs during embryogenesis. One such function could be to toggle accessibility of TE-derived enhancers, resulting in the regulation of nearby genes in a cell-type specific fashion. In this regard, the arms race between KRAB-ZFPs and TEs could be an important source of innovation for gene regulation.
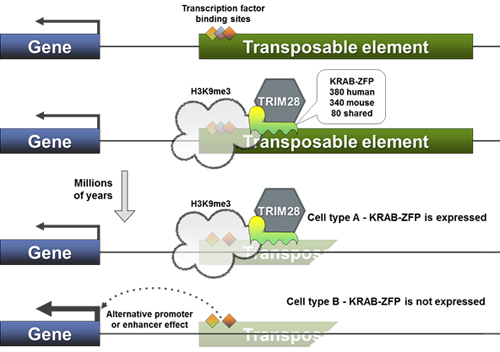
So far, only a handful of the 350 human KRAB-ZFPs have been functionally characterized, yet many of those few have important and novel biological functions, such as ZFP57 and imprinting, or PRDM9 and the regulation of meiotic recombination. Moreover, studies of their common effector TRIM28 suggest potential roles for KRAB-ZFPs in the latency cycle of disease-causing viruses (CMV, KSHV), the regulation of cellular development and the neogenesis of cancer.
Through a mix of bioinformatics and molecular biology, our lab is set to elucidate the biological functions of some of the most evolutionary conserved KRAB-ZFPs. We are also interested in the characterization of their peculiar evolution dynamics, especially in the current human population. The ever-increasing numbers of full human genomes being sequenced make it possible to get a deeper look at the contribution of KRAB-ZFPs and transposable elements to disease risk factors and other complex traits.
