Submitted by Christine Alexander on Fri, 26/10/2018 - 13:51
Differences in traits between individuals are predominantly genetically conferred. However, evidence from human population studies and experiments in animal models suggests that environmentally induced phenotypic changes might be transmitted across generations via non-genetic mechanisms. This has important implications for health and disease and has captured the attention of society at large, with a growing concern that lifestyle choices can have a lasting legacy across generations.
Transposable elements are repetitive DNA sequences that make up almost half of the mammalian genome. One compelling hypothesis, underpinned by experimental evidence in the mouse, suggests that transposable elements might mediate non-genetic inheritance through epigenetic mechanisms (i.e. chemical modifications to our genetic material that can influence gene function without changing the DNA sequence itself).
While most transposable elements are epigenetically silenced, previous data from two mouse models indicates that differences in appearance between genetically identical mice could indeed be conferred by variable epigenetic states at specific transposable elements, and that these differences in appearance could be passed on to the next generation. This has made these mouse models celebrated paradigms of so-called epigenetic inheritance in mammals.
The Ferguson-Smith Group in the Department of Genetics was interested in determining the genome-wide prevalence of this phenomenon and set out to identify epigenetically variable transposable elements across the mouse genome. The product of this research was published in Cell this week [1]. The article, co-authored by Anastasiya Kazachenka and Tessa Bertozzi, describes the identification of such regions and explores their genomic and epigenomic features.
The work shows that only a subset of epigenetically variable elements can influence neighbouring gene expression, but most are enriched in CTCF, a transcription factor involved in the regulation of genomic architecture. This opens up the possibility that these elements may regulate genes in a long-range capacity.
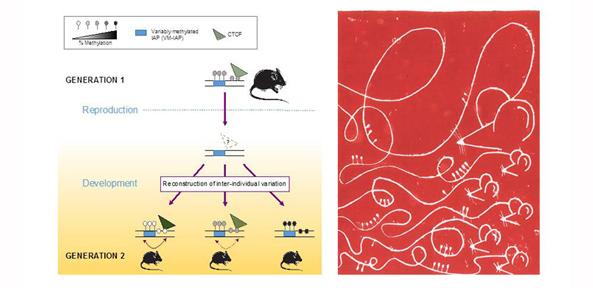
Importantly, the study asked whether there is a memory of parental epigenetic states in the next generation. Breeding intensive experiments found only one example where the maternal epigenetic state was reflected in the offspring, but the effect size was relatively minor. Instead, most of the studied regions show complete re-construction of epigenetic variability after passage through the germline regardless of parental epigenetic state. It is therefore unlikely that substantial epigenetic inheritance is occurring at these regions.
This body of work provides a repertoire of loci to study the effects of variable epigenetic states on genome function and suggests that epigenetic inheritance across generations is less common than people might think.
>> Tessa Bertozzi Naked Scientist interview [print]
>> Tessa Bertozzi Naked Scientist interview [link to podcast]
[1] Kazachenka, A., Bertozzi, T.M., Sjoberg-Herrera, M.K., Walker, N., Gardner, J., Gunning, R., Pahita, E., Adams, S., Adams, D., Ferguson-Smith, A.C., Identification, Characterization, and Heritability of Murine Metastable Epialleles: Implications for Non-genetic Inheritance, Cell (2018), https://doi.org/10.1016/j.cell.2018.09.043
The image above and on the home page displays on the left the graphical abstract of the research, and on the right a linocut print made by co-author Tessa Bertozzi to give an artistic representation of our study. The linocut print depicts a mouse’s DNA and that of its offspring, illustrating the varying levels of methyl groups observed across individuals at the regions identified in the study.
